Dr. Yi Xu recently successfully defended her thesis titled “Towards Automated Epigenetics: Sample Processing with Droplet Microfluidics."
She will be heading to Zymo Research to continue her successful career. Congratulations Yi!
Dr. Yi Xu recently successfully defended her thesis titled “Towards Automated Epigenetics: Sample Processing with Droplet Microfluidics."
She will be heading to Zymo Research to continue her successful career. Congratulations Yi!

Recently, various members of the Bailey lab showed off their hard work. At the University's Karle Symposium, Colleen picked up an award for her oral presentation, "Microfluidic Platform for Rapidly Incorporating Total Membrane Protein Content from Whole Cell Lysate into Nanodisc Libraries Enables Activity-Based Profiling." Cole also talked his poster about "Cytokine Profiling for the Diagnosis of Chorioamnionitis Using Silicon Photonic Microring Resonator Arrays" up for an award.
Heather also just got back from the AACC Annual Scientific Meeting & Clinical Lab Expo where she presented her work, "A Machine Learning Approach to Inflammatory Cytokine Profiling Reveals Diagnostic Signatures for Latent Tuberculosis Infection and Reactivation Risk Stratification," and earned the AACC Student Poster Contest, Second Place and Personalized Medicine Division Outstanding Abstract Award!
Way to go, everyone!
ABSTRACT
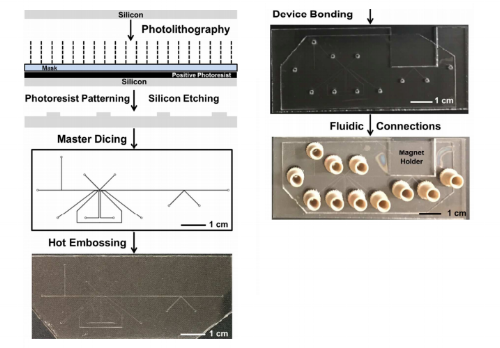
We have developed droplet microfluidic devices in thermoplastics and demonstrated the integration of key functional components that not only facilitate droplet generation, but also include electric field-assisted reagent injection, droplet splitting, and magnetic field-assisted bead extraction. We manufactured devices in poly(methyl methacrylate) and cyclic olefin polymer using a hot-embossing procedure employing silicon masters fabricated via photolithography and deep reactive ion etching techniques. Device characterization showed robust fabrication with uniform feature transfer and good embossing yield. Channel modification with heptadecafluoro-1,1,2,2-tetrahydrodecyltrichlorosilane increased device hydrophobicity, allowing stable generation of 330-pL aqueous droplets using T-junction configuration. Picoinjector and K-channel motifs were also both successfully integrated into the thermoplastic devices, allowing for robust control over electric field-assisted reagent injection, as well as droplet splitting with the K-channel. A magnetic field was also introduced to the K-channel geometry to allow for selective concentration of magnetic beads while decanting waste volume through droplet splitting. To show the ability to link multiple, modular features in a single thermoplastic device, we integrated droplet generation, reagent injection, and magnetic field-assisted droplet splitting on a single device, realizing a magnetic bead washing scheme to selectively exchange the fluid composition around the magnetic particles, analogous to the washing steps in many common biochemical assays. Finally, integrated devices were used to perform a proof-of-concept in-droplet β-galactosidase enzymatic assay combining enzyme-magnetic bead containing droplet generation, resorufin-β-D-galactopyranoside substrate injection, enzyme-substrate reaction, and enzyme-magnetic bead washing. By integrating multiple droplet operations and actuation forces we have demonstrated the potential of thermoplastic droplet microfluidic devices for complex (bio)chemical analysis, and we envision a path toward mass fabrication of droplet microfluidic devices for a range of (bio)chemical applications.
Click here to read the manuscript!

ABSTRACT
The first step in chromatin-based epigenetic assays involves the fragmentation of chromatin to facilitate precise genomic localization of the associated DNA. Here, we report the development of a droplet microfluidic device that can rapidly and efficiently digest chromatin into single nucleosomes starting from whole-cell input material offering simplified and automated processing compared to conventional manual preparation. We demonstrate the digestion of chromatin from 2500–125 000 Jurkat cells using micrococcal nuclease for enzymatic processing. We show that the yield of mononucleosomal DNA can be optimized by controlling enzyme concentration and incubation time, with resulting mononucleosome yields exceeding 80%. Bioinformatic analysis of sequenced mononucleosomal DNA (MNase-seq) indicated a high degree of reproducibility and concordance (97–99%) compared with conventionally processed preparations. Our results demonstrate the feasibility of robust and automated nucleosome preparation using a droplet microfluidic platform for nucleosome positioning and downstream epigenomic assays.
Click here to read the manuscript!

The award recognizes research and other contributions in advancing the field of microchemistry. You can read more about the award and Professor Bailey's accomplishments here!

ABSTRACT
Analysis methods based upon the quantitative, real-time polymerase chain reaction are extremely powerful; however, they face intrinsic limitations in terms of target multiplexing. In contrast, silicon photonic microring resonators represent a modularly multiplexable sensor array technology that is well-suited to the analysis of targeted biomarker panels. In this manuscript we employ an asymmetric polymerase chain reaction approach to selectively amplify copies of cDNAs generated from targeted miRNAs before multiplexed, label-free quantitation through hybridization to microring resonator arrays pre-functionalized with capture sequences. This method, which shows applicability to low input amounts and a large dynamic range, was demonstrated for the simultaneous detection of eight microRNA targets from twenty primary brain tumor samples with expression profiles in good agreement with literature precedent. Click here to read the manuscript.
The group just got back from a breather down in Florida. When not giving talks or presenting their posters, everyone soaked up the sun and spent a night out at Universal Studios.

A method for quantifying biologically relevant long-non-coding RNAs by combining nucleic acid amplification via asymmetric polymerase chain reaction (PCR) with label-free PCR product detection using silicon photonic microring resonator arrays is described. This approach eliminates the need for fluorophores, which presents a limit for spectral multiplexing in conventional qPCR methods, and rather offers potential for much higher levels of plexity by spatially arraying capture probes. Here, we demonstrate the potential of this technique to detect two differentially expressed lncRNA transcripts and an internal control mRNA transcript in different commercial human tissue specimens, as well as in a glioblastoma cell line using only nanogram input amounts of total RNA. The obtained results were validated using single-plex RT-qPCR and found to be in good agreement, demonstrating the potential of this technique for lncRNA quantification applications. Click here to read the manuscript.

Dr. Heather Robison recently successfully defended her thesis titled “Multiplexed Immunoassay Development for Precision Medicine Diagnostics and Protein Characterization using Silicon Photonic Microring Resonators".
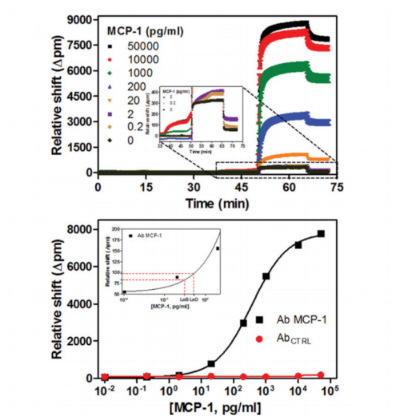
Whispering gallery mode (WGM) sensors are a class of powerful analytical techniques defined by the measurement of changes in the local refractive index at or near the sensor surface. When functionalized with target-specific capture agents, analyte binding can be measured with very low limits of detection. There are many geometric manifestations of WGM sensors, with chip-integrated silicon photonic devices first commercialized because of the robust, wafer-scale device fabrication, facile optical interrogation, and amenability to the creation of multiplexed sensor arrays. Using these arrays, a number of biomolecular targets have been detected in both label-free and label-enhanced assay formats. For example, sub-picomolar detection limits for multiple cytokines were achieved using an enzymatically enhanced sandwich immunoassay that showed high analyte specificity suitable for detection in complex, clinical matrices. This protocol describes a generalizable approach for the development of quantitative, multiplexed immunoassays using silicon photonic microrings as an example WGM platform.